DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation | Genome Biology | Full Text

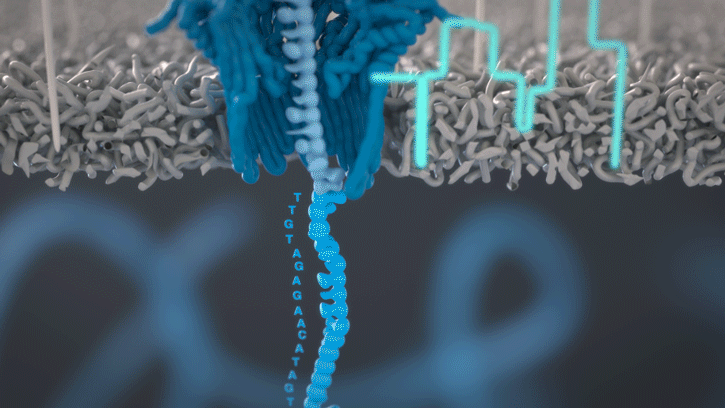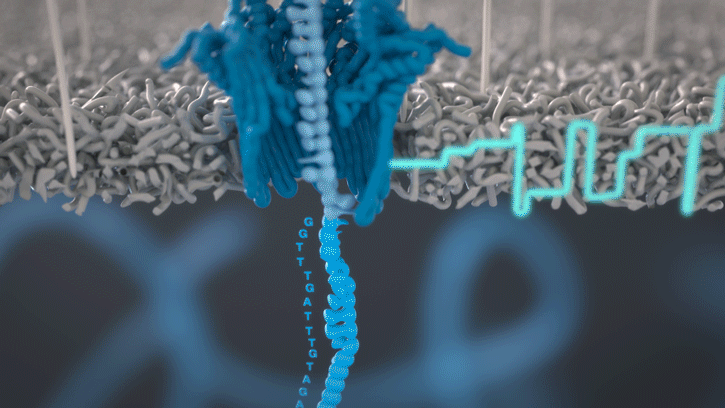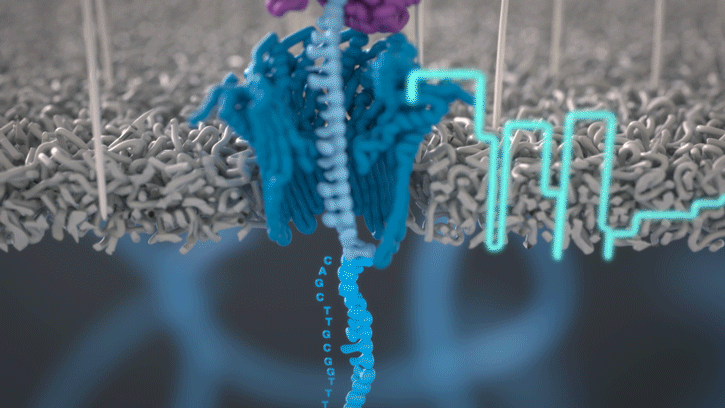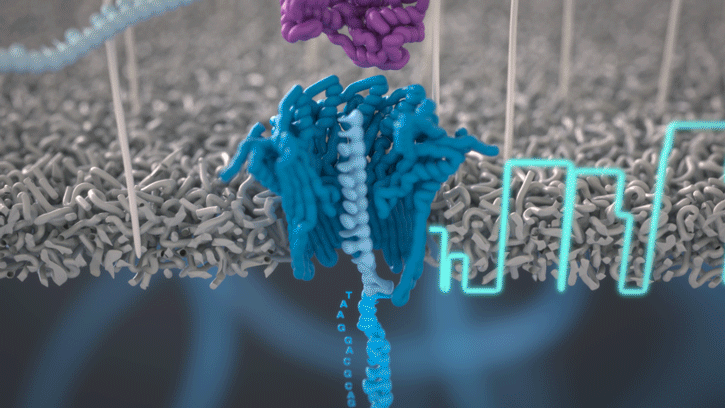
Oxford Nanopore on Twitter: "CB: @nanopore has an information-rich signal which can be deconvoluted in real time. Basecalling updates have driven considerable improvements over the past two years. https://t.co/US1mt94Wub" / X
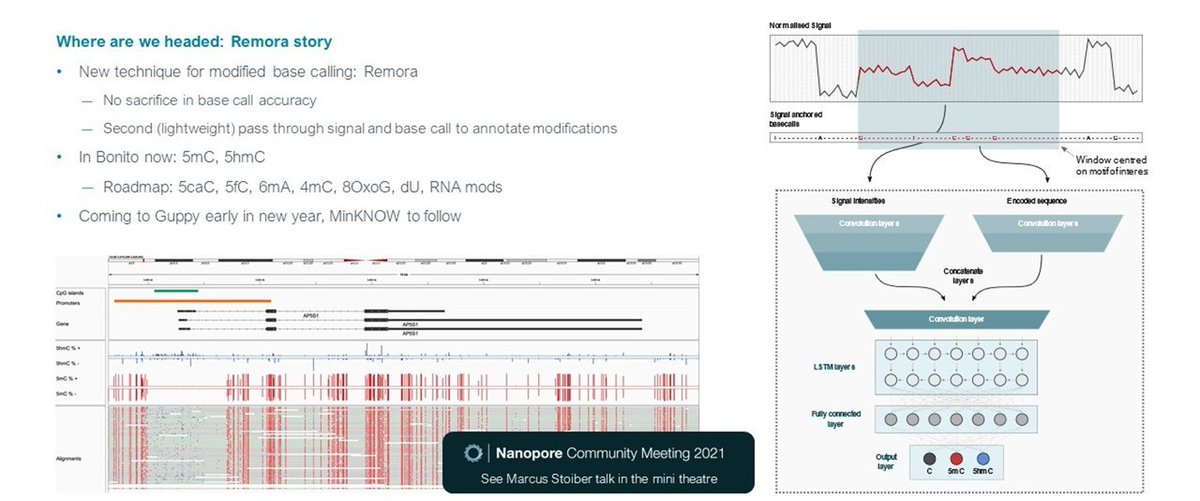
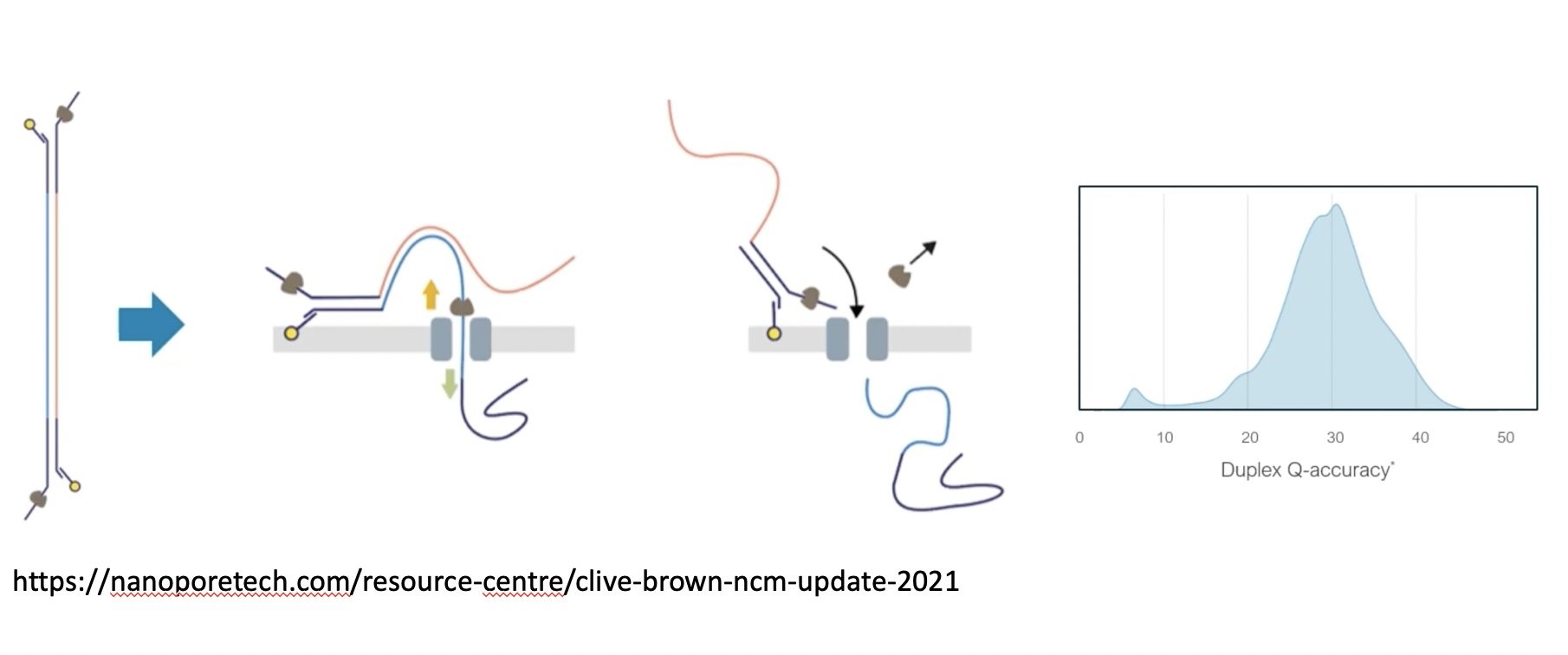
Oxford Nanopore on Twitter: "SR: Remora is 2nd, lightweight pass through the signal, that can run with basecalling, sacrificing no accuracy when delivering methylation analysis in the same experiment. Now in Bonito

Frontiers | MasterOfPores: A Workflow for the Analysis of Oxford Nanopore Direct RNA Sequencing Datasets
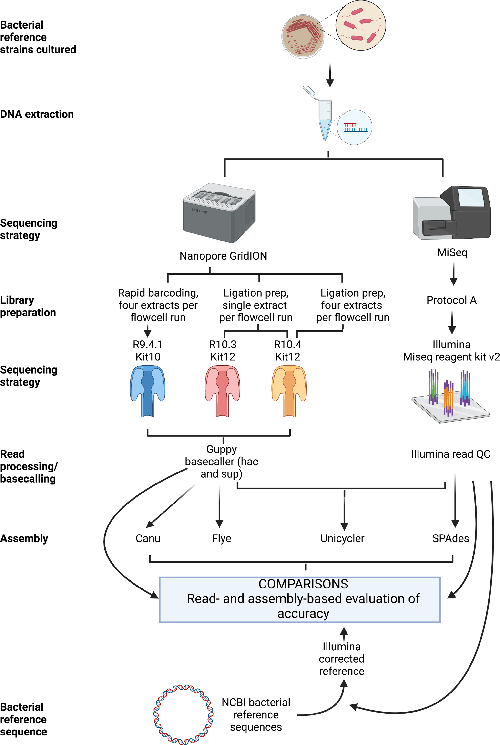
Comparison of R9.4.1/Kit10 and R10/Kit12 Oxford Nanopore flowcells and chemistries in bacterial genome reconstruction | Microbiology Society
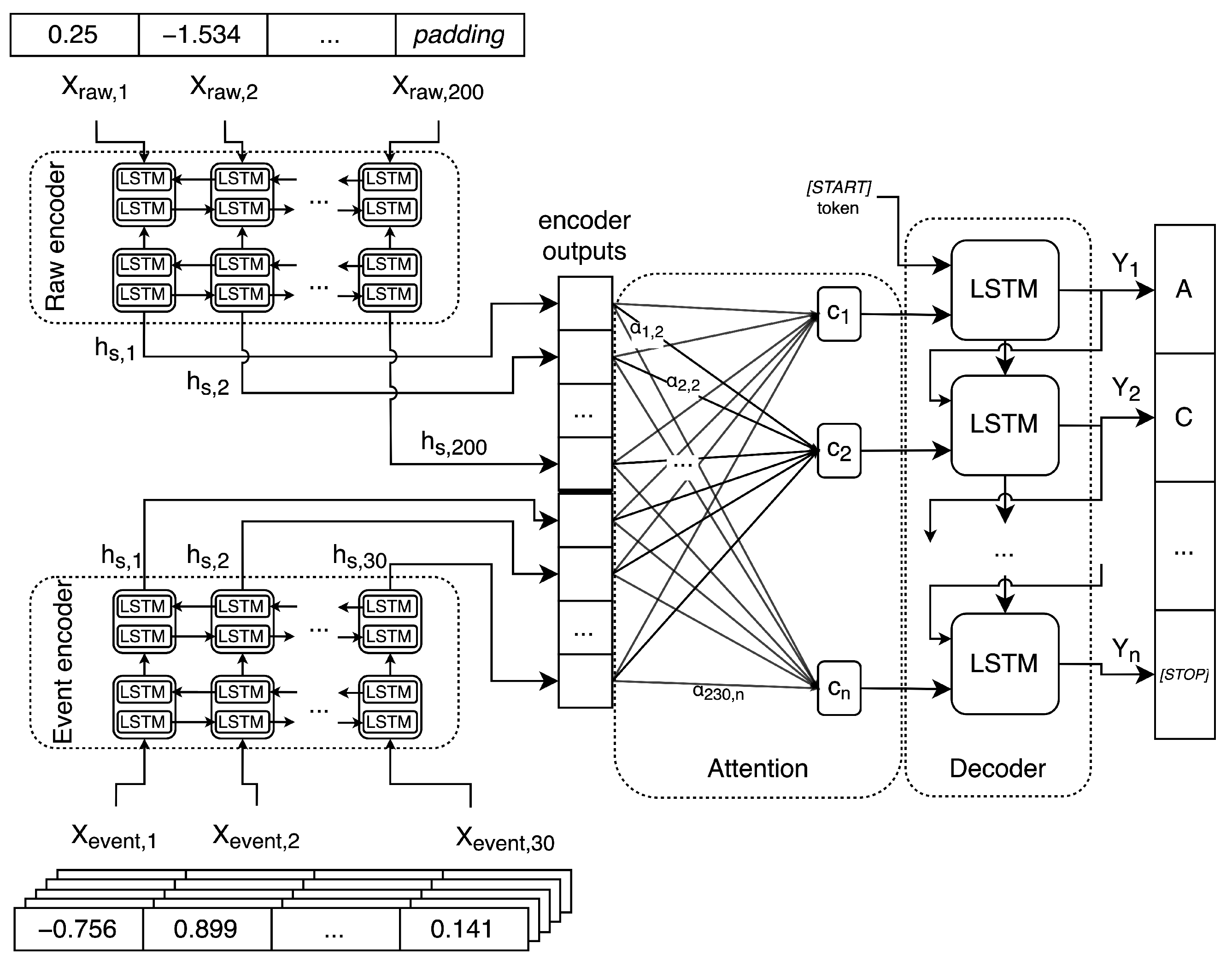
Sensors | Free Full-Text | Basecalling Using Joint Raw and Event Nanopore Data Sequence-to-Sequence Processing
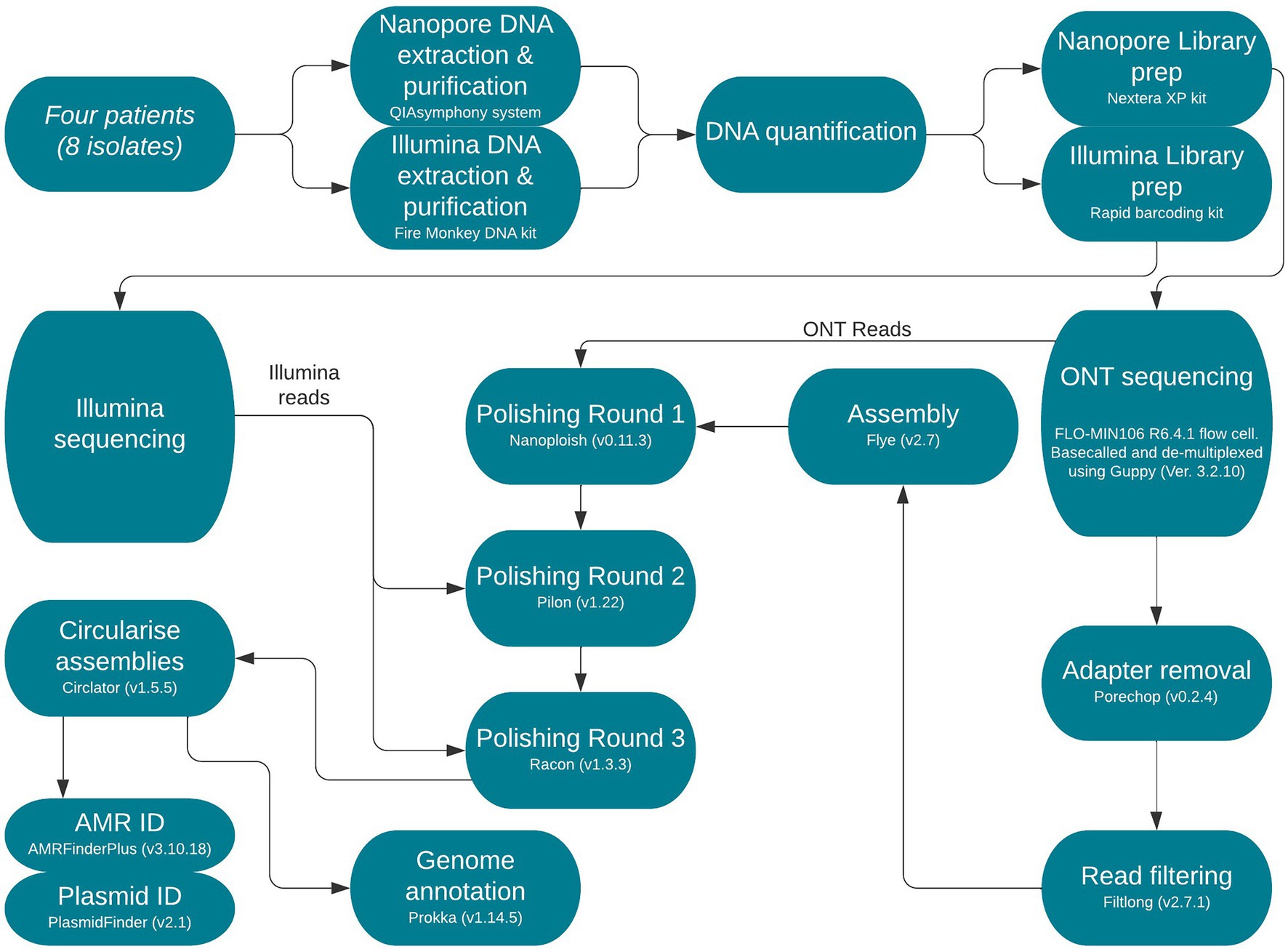
Frontiers | Use of Nanopore Sequencing to Characterise the Genomic Architecture of Mobile Genetic Elements Encoding blaCTX-M-15 in Escherichia coli Causing Travellers' Diarrhoea

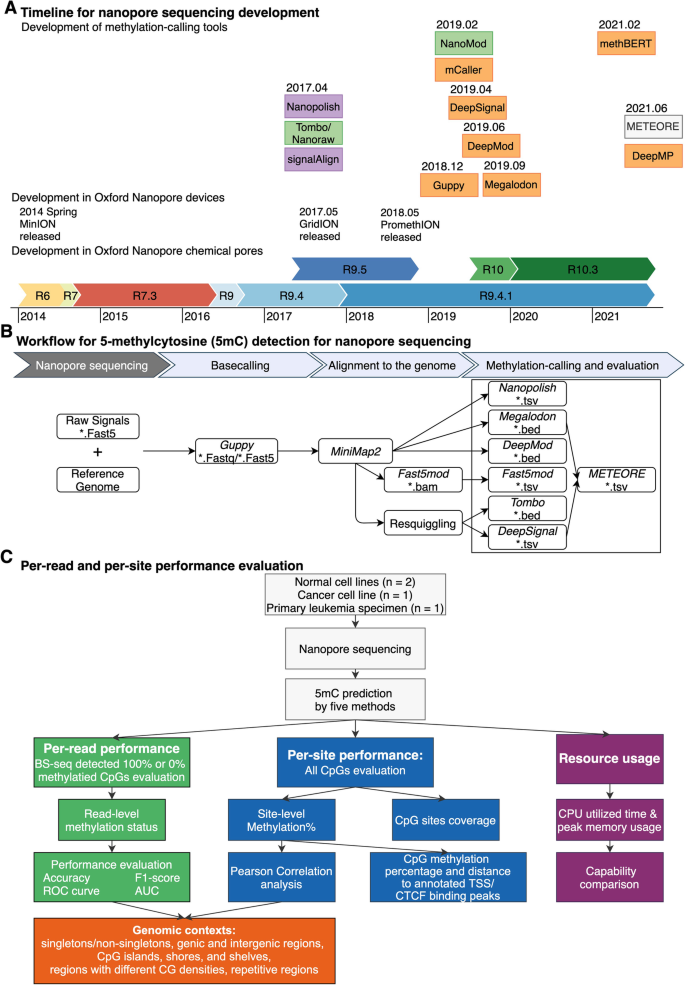
Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing | Nature Communications
GitHub - asadprodhan/GPU-accelerated-guppy-basecalling: GPU-accelerated guppy basecalling and demultiplexing on Linux
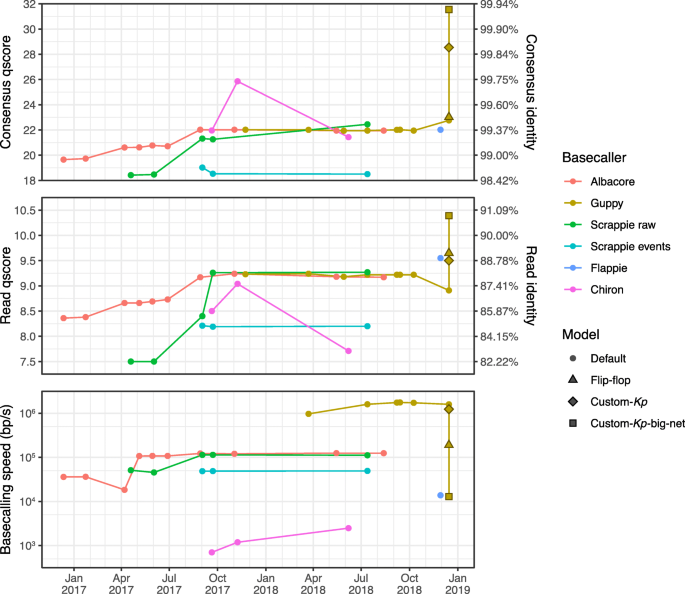
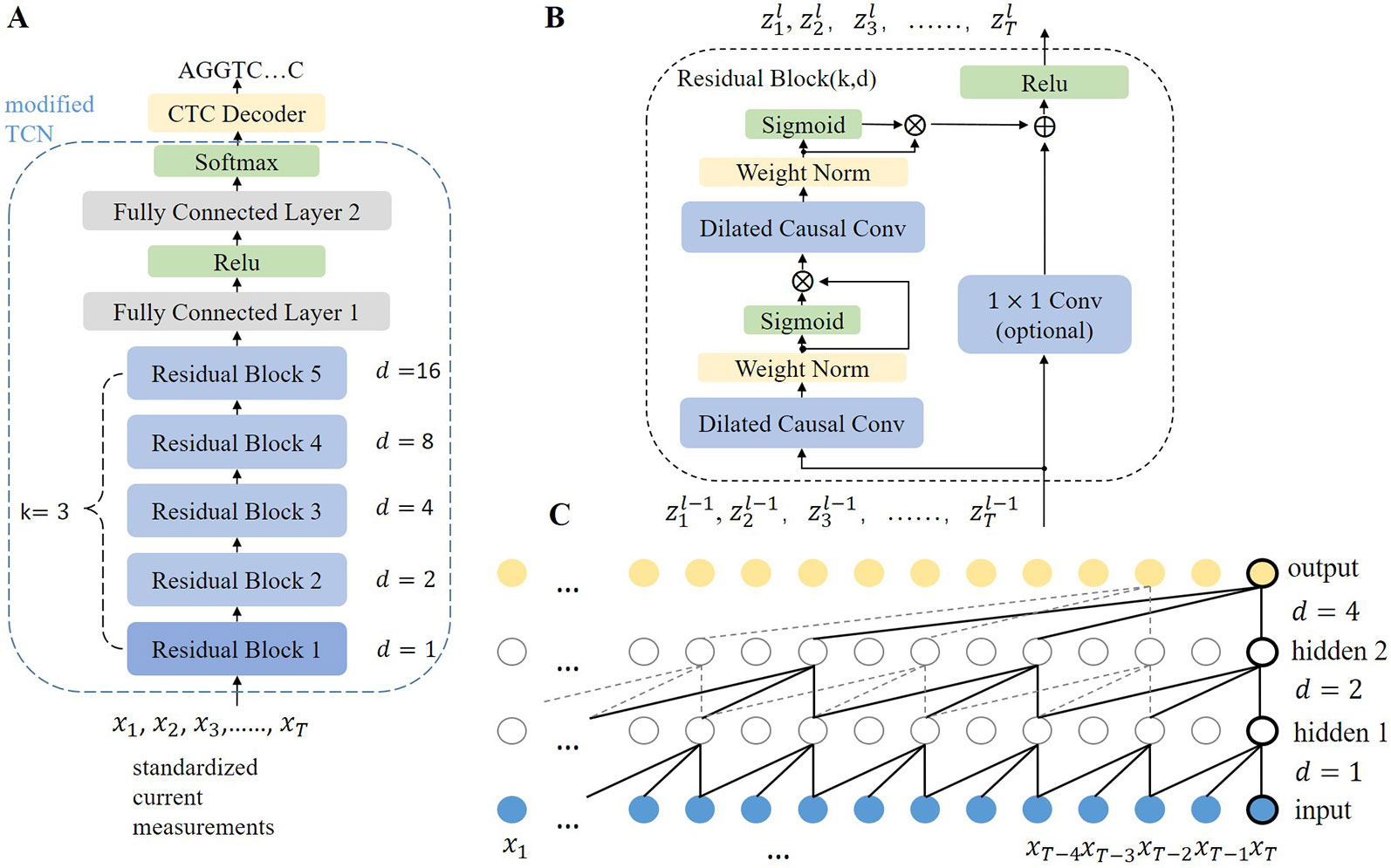
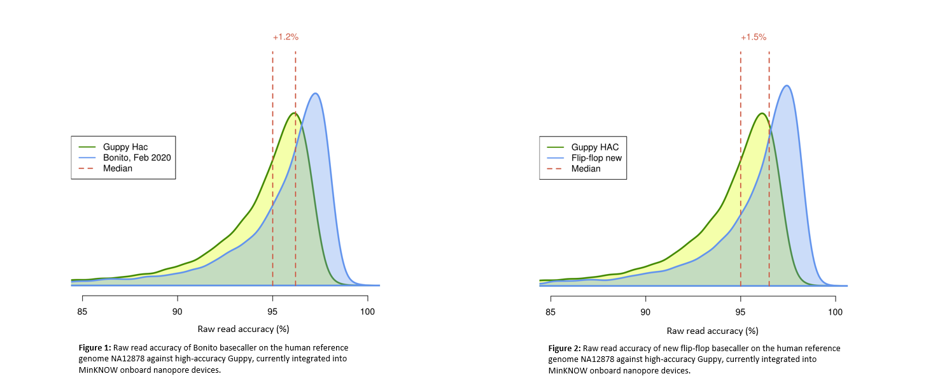
Performance of neural network basecalling tools for Oxford Nanopore sequencing | Genome Biology | Full Text

Consensus decoding improves sequencing accuracy. Reads were run through... | Download Scientific Diagram